Access paper: https://doi.org/10.1101/2020.10.02.322529
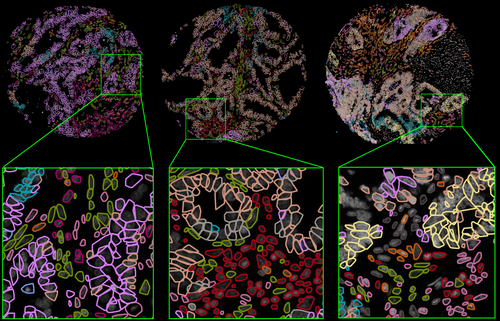
This paper shows how to build a data-driven and computationally unbiased phenotypic hierarchy of cell types, cell states, and their spatial configurations from multi to hyperplexed tissue samples to characterize spatial intratumor heterogeneity, discover recurrence-associated microdomains, and harness spatial network biology for precision medicine applications.
This work in combination with our Nature Communications study completes the development of our spatial analytics and xAI-based early discovery and development software analytics platform and service, TumorMapr™.
With this advancement, TumorMapr can provide mechanistic insights into disease biology such as the generation of outcome-specific new pathways, new cellular phenotypes, and iterative experimental testing of additional biomarkers on the discovered microdomains.